Fundamental research into DNA “bubbles” will help scientists understand how genetic programmes unfold
Posted on Tuesday 28 January 2025
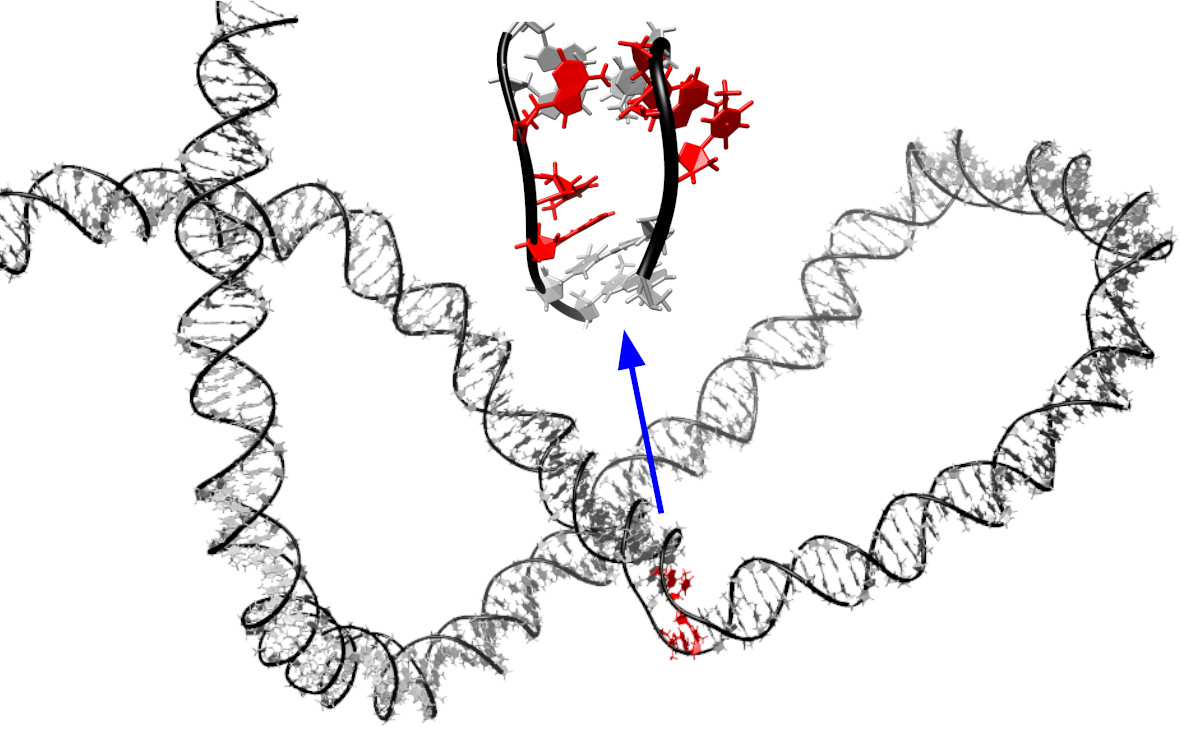
These findings mean that genetic information is easier to read than was previously thought, enhancing scientists’ abilities to predict which hereditary information will be accessed first in the pursuit of genetic engineering and disease detection.
Since the discovery of the double helix, scientists have known that the molecular parts of DNA that carry genetic information - the four nucleobases A, T, C, and G - are inside the structure, which makes them hard to get to. Previously, it was believed that specialised proteins were required to generate these “bubbles”. This current study indicates that they are mostly programmed in the DNA sequence (AT-rich sequences being weaker than GC-rich ones).
This means that really simple models can be used to guess which parts of genomes will be read first, helping scientists to learn more about how genetic diseases develop and how to tune genes for genetic engineering.
Dr Agnes Noy, Senior Lecturer in the School of Physics, Engineering and Technology, who led the research, said: “Our results should help other scientists to grasp how the genetic information contained on genomes unfolds, including during disease.”
These results were already suggested in different types of experiments (2). However, by performing computational simulations, Dr Noy’s research provides us with a thorough view of the conditions under which DNA bubbles, connecting data and models from very different sources to construct a holistic image of the process.
View an extract from a simulation showing the formation of a "bubble", by Dr Agnes Noy.
(1) M Burman and A Noy (2025) “Atomic description of the reciprocal action between supercoils and melting bubbles on linear DNA” Paper accepted in Phys Rev Lett
(2) J. M. Fogg, A. K. Judge, E. Stricker, H. L. Chan, andL. Zechiedrich (2021) “Supercoiling and looping promote DNA base accessibility and coordination among distant sites” Nat. Commun. 12, 5683.
See further coverage of this research story published by phys.org.
